MolecularFeaST
MolecularFeaST
The Molecular Feature Selection Tool (MolecularFeaST) is a free-to-use, standalone application that streamlines the use of our feature selection algorithm. MolecularFeaST has been used to find biomarkers based on numerous -omics profiles including: RNA-seq, small RNA-seq, RT-qPCR data, NanoString profiling, microarray data, srRNA expression, bisulfite (DNA methylation) sequencing, and protein mass spectroscopy. Contact us to request a copy of the software.
Discover biomarkers in your data
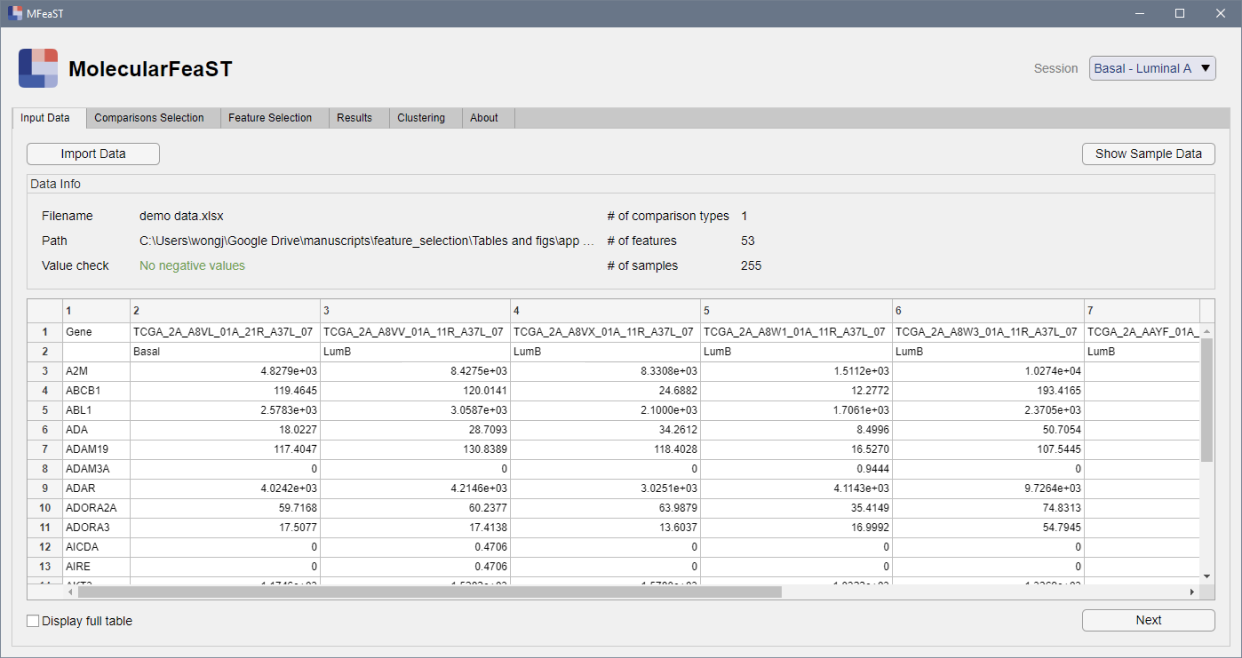
Import dataset
Import -omics profiles from Excel or .csv files with a single click.
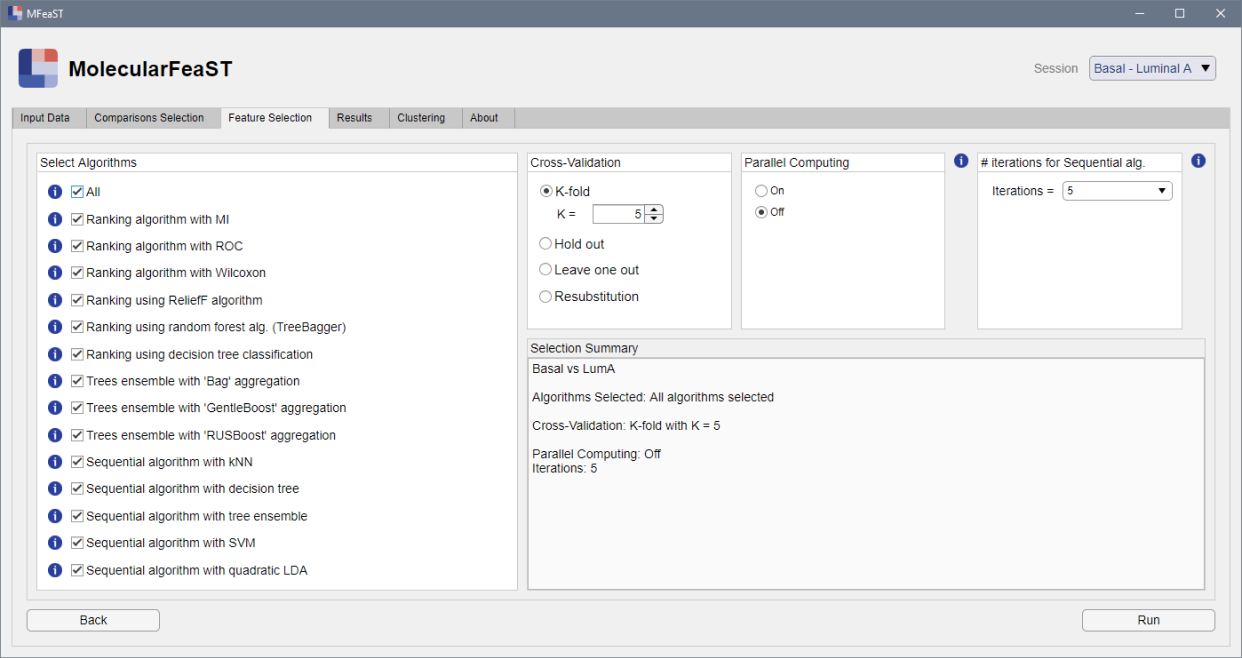
Choose comparisons and feature selection algorithms
Choose the classes you wish to discriminate, and which feature selection algorithms you would like to apply. Choose any number of the 14 available algorithms.
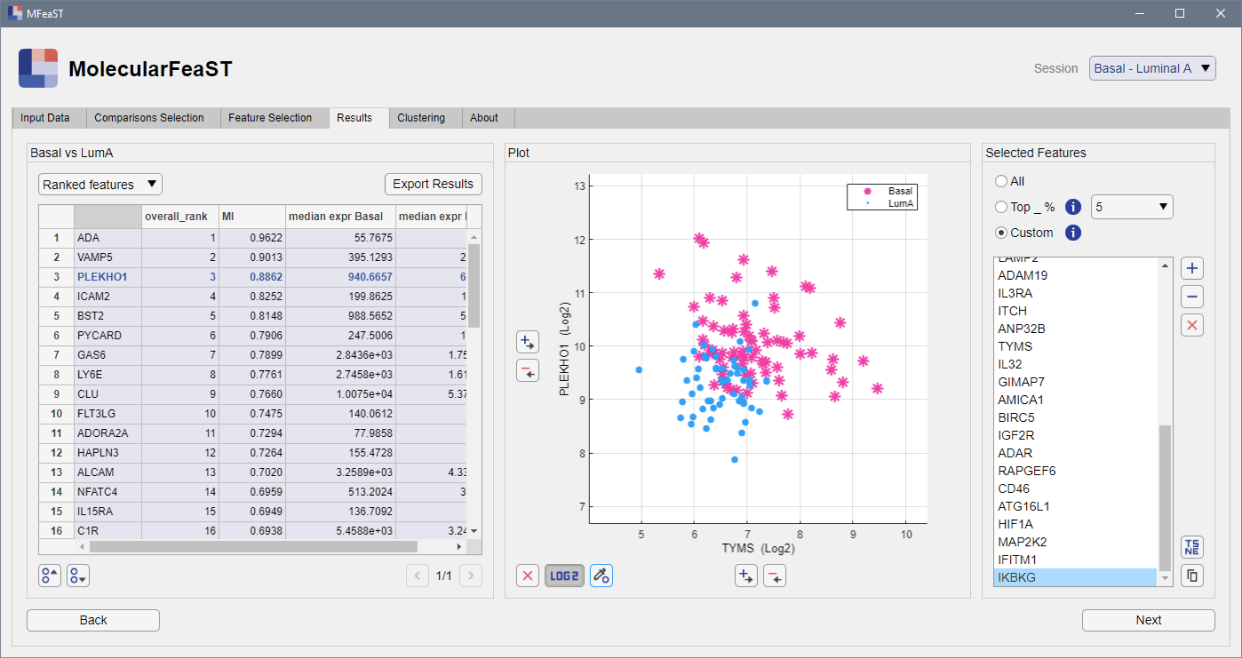
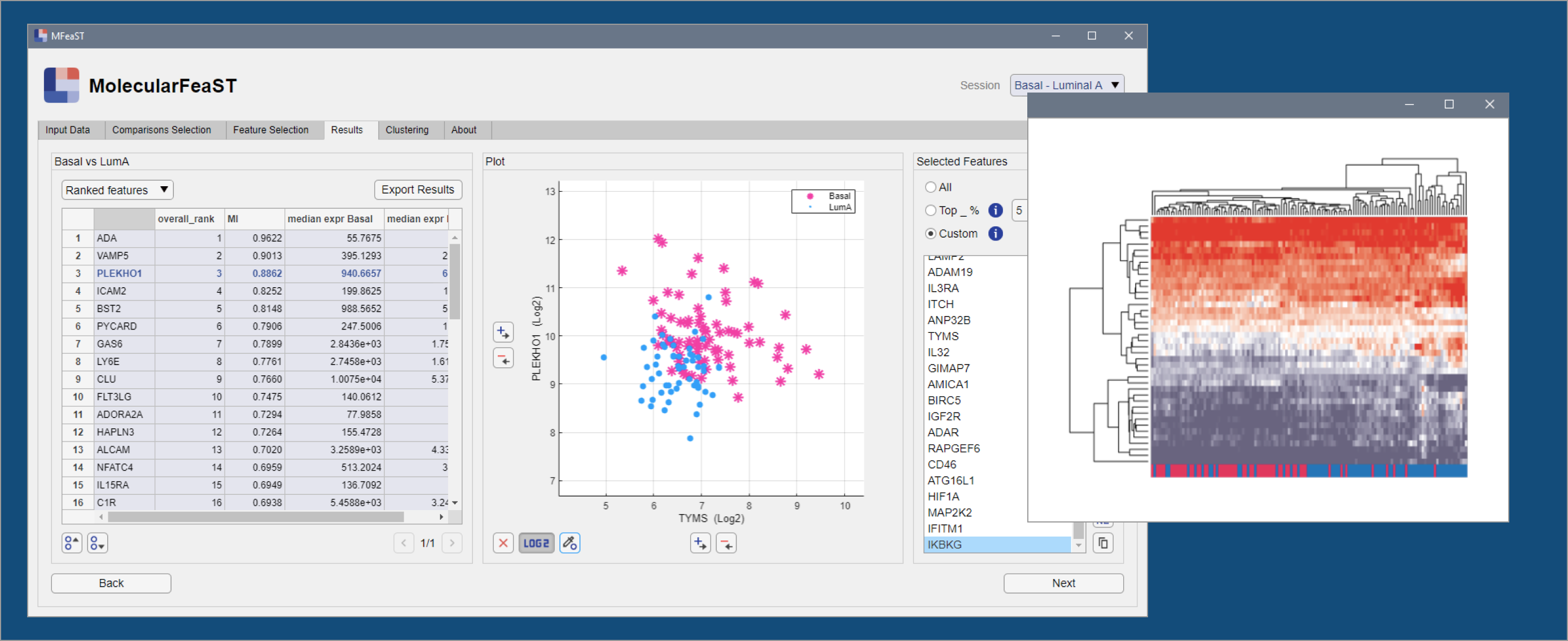
View results and select desired features
Use the scatterplot window to view the ranked features and select candidate biomarkers. Select manually or by top percentile.
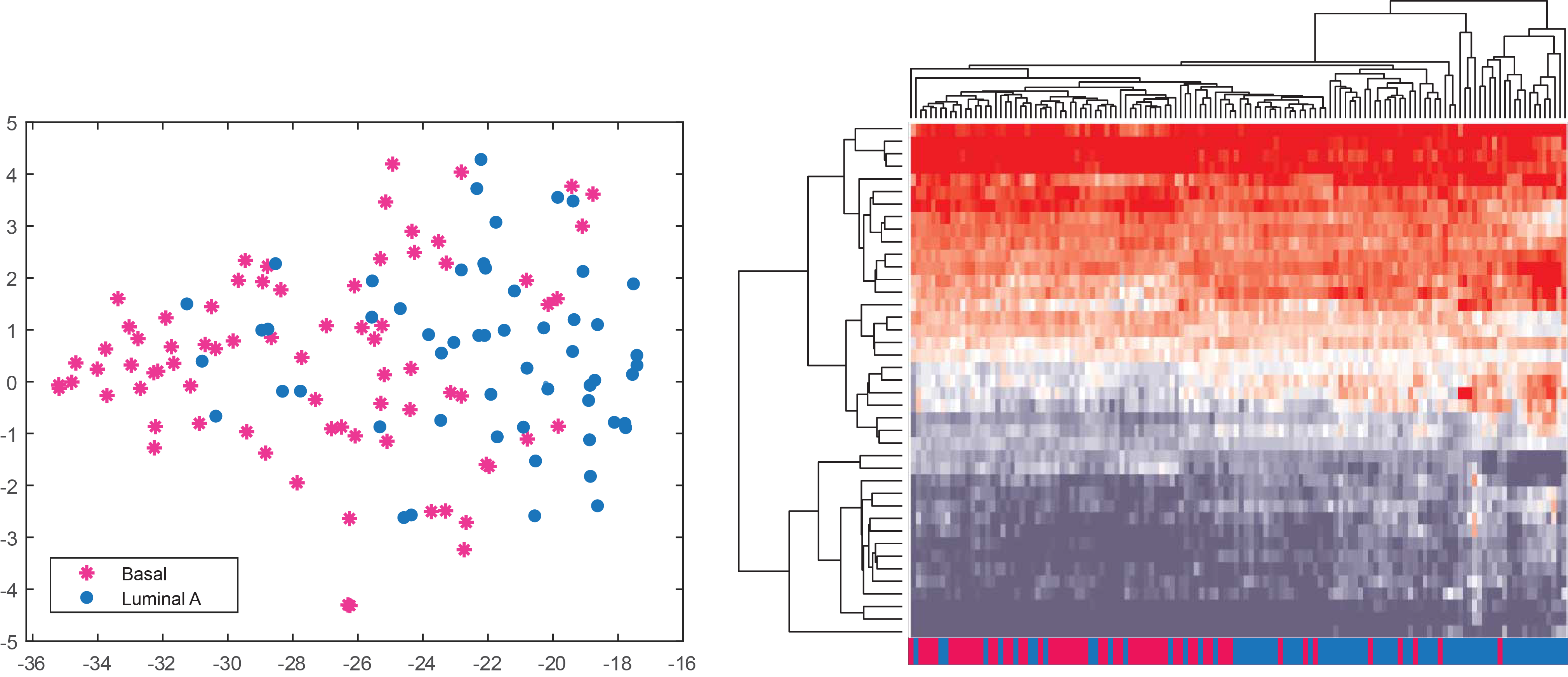
Generate and save clustering analyses
Create t-SNE and heatmap visualizations from your selected features.
System Requirements
| Specification | Minimum Requirement | Recommended Requirement |
|---|---|---|
| Operating System | Windows 7 SP 1 or macOS 10.14 |
Windows 7 SP 1 and later or macOS 10.14 and later. The software must be installed as an administrator |
| Disk Space | 10GB | 15GB or more |
| RAM | 4GB | 8GB or more |
| CPU | Any Intel or AMD x86-64 processor | Any Intel or AMD x86-64 processor with four logical cores or more and AVX2 instruction set support |
Citation
If you use MolecularFeaST, please cite the following:
Gerolami, J.; Wong, J.J.M.; Zhang, R.; Chen, T.; Imtiaz, T.; Smith, M.; Jamaspishvili, T.; Koti, M.; Glasgow, J.I.; Mousavi, P.; Renwick, N.; and Tyryshkin, K. A Computational Approach to Identification of Candidate Biomarkers in High-Dimensional Molecular Data. Diagnostics 2022, 12,1997.